
Marine
Microbial
Ecology
Investigating the hidden engines of our oceans
Our Team
We are curious and dedicated scientists exploring how microbial diversity, dynamics, and interactions impacts our ocean and climate.
Our group strives to foster a diverse, equitable, and inclusive research environment that celebrates diversity as a moral value. We recognize that diverse backgrounds, different life experiences, and global perspectives are essential to research excellence. We aspire to a stimulating, collaborative, and open research environment, inspired by our individual and shared passions and curiosities about nature.
Sea-surface microlayer biodiversity, activity, and interactions
Investigating the impact of ocean alkalinity enhancement on nitrification
Microbe-to-microbe interactions using cultivation-independent methods
The impact & implications of extreme conditions on marine microbial interactions
Bio- & ecological controls on marine nitrification with an archaeal focus
Seasonality of carbon fixation and nitrification rates in the Baltic Sea
Jana Hinz
.jpg)
Germany
Bachelor Student
Time-series sampling Pro
Lara Schroeder
.jpg)
Germany
Bachelor Student
Seasonality of phages in Southwest Baltic Sea
Alumni
Jana Hinz
.jpg)
Germany
Bachelor Student
Lara Schroeder
.jpg)
Germany
Bachelor Student
Seasonality of phages in Southwest Baltic Sea
Add paragraph text. Click “Edit Text” to update the font, size and more. To change and reuse text themes, go to Site Styles.
We are curious and dedicated scientists exploring how microbial diversity, dynamics, and interactions impacts our ocean and climate.
Our group strives to foster a diverse, equitable, and inclusive research environment that celebrates diversity as a moral value. We recognize that diverse backgrounds, different life experiences, and global perspectives are essential to research excellence. We aspire to a stimulating, collaborative, and open research environment, inspired by our individual and shared passions and curiosities about nature.
Jana Hinz
.jpg)
Germany
Bachelor Student
Time-series sampling Pro
Lara Schroeder
.jpg)
Germany
Bachelor Student
Seasonality of phages in Southwest Baltic Sea
Research
Biodiversity and interactions in the plankton
Our research addresses a broad question: How do microbial eukaryotes, bacteria, archaea, and viruses interact to impact ocean ecosystem function, productivity, and climate?
Integrating across trophic levels and the tree of life, we apply a variety of methods
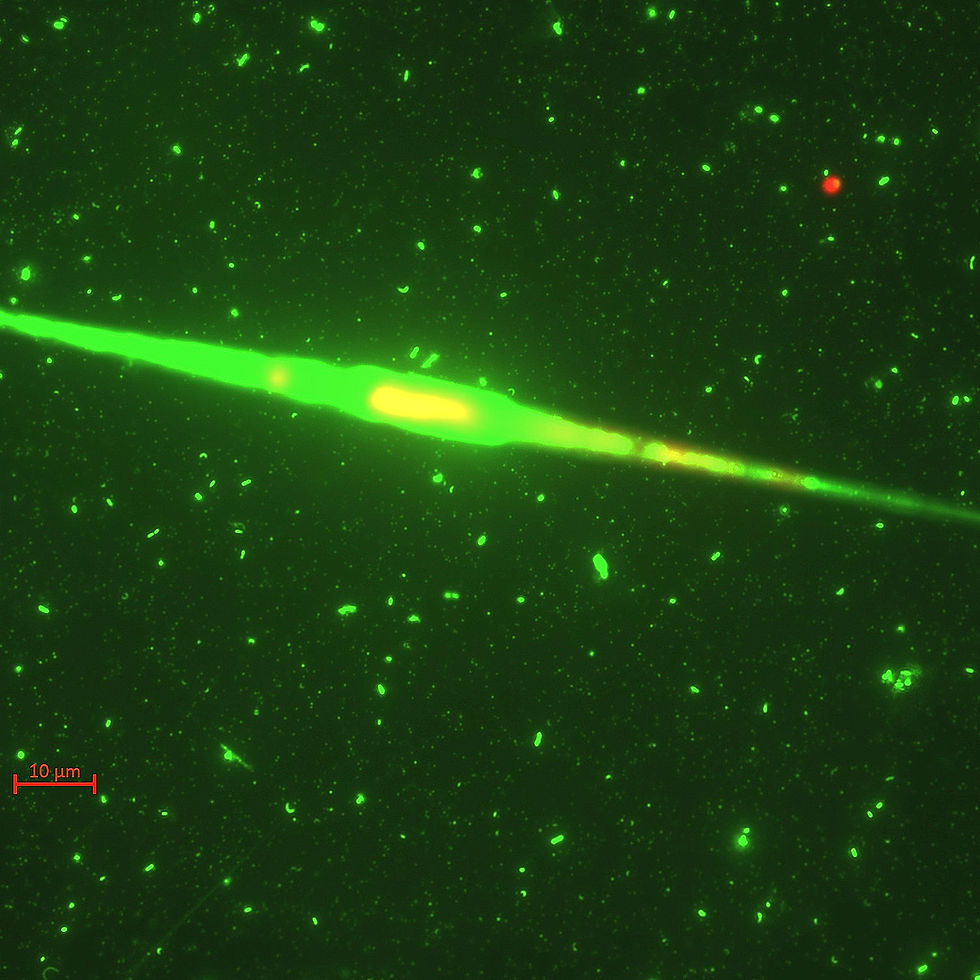
Typical SYBR green fluorescence microscopy image of marine microbes with a large diatom, alongside bacteria and many viruses
From single cell to global scale, we use tools of molecular ecology and meta-omic technologies to discover novel micro-organisms, and their ecology, and function. One of the workhorses we use in the lab is rRNA gene sequencing, whereby we use universal primers to amplify and sequence eukaryotes, bacteria, and archaea, and chloroplasts to examine dynamics across all three domains. We also use high-throughput library generation for Illumina as well as innovations toward miniaturized extraction and library workflows for microliter samples and small experimental units to increase throughput and access micro-environments, while maintaining accuracy. For single cell sequencing we utilize flow cytometry single cell sorting of prokaryotes and eukaryotes as well as droplet-based approaches.
Computationally, we use state-of-the-art methods for analyzing rRNA gene sequences as well as genomics, taking advantage of the University of Kiel’s HPC computing cluster. We also teach bioinformatics in the University of Kiel’s Biological Oceanography Master’s program.
Recent Examples: Calayag et al., 2025 and Ghotbi et al. Pre-print
Video demonstrates microfluidic water microdroplet creation from Zheng et al., 2022.

We focus on isolating and experimenting with environmental-relevant but often fastidiously growing microorganisms from marine environments to gain insights into their physiology, metabolic pathways, and ecological roles within microbial communities. We target fastidious prokaryotes, especially ammonia oxidizing archaea, which grow slowly and are overlooked in standard surveys. These can use these microbes to serve as models for studying interactions, physiology, adaptation to extreme conditions like warming temperatures, or changing pH such as via ocean alkalinity enhancement.
Image at right, nitrite colorometric assay in a microwell plate demonstrating growth of ammonia oxidizing archaea.
Recent Example: Kim et al. Pre-print

To quantify microbial activity and material exchange, we conduct tracer incubations using stable isotopes such as 13/14C and 15N, tracking their incorporation into biomass and metabolites over time. We are especially using these tools to track the rates and top-down controls on nitrification in the ocean. We apply these methods to both lab-based microcosms and field samples from oceanographic expeditions, helping to disentangle the contributions of individual species in microbial networks.
These tools are important components of our German Research Foundation (DFG) funded proposal: N-control, Biological, ecological, and evolutionary controls on marine ammonia oxidation.

We construct ecological networks by integrating high-resolution environmental datasets, such as from amplicon sequencing and metagenomics. This allows us to model species interactions, including competition, mutualism, and predation, across spatial and temporal scales in marine ecosystems. Our teaching modules in the Biological Oceanography Master’s program have included hands-on training in network visualization tools like Cytoscape.
Recent Example: Calayag et al., 2025

Leveraging meta-omics datasets from unique sampling sites, such as Arctic Ocean and anaerobic munition dump sites in the Baltic Sea, we conduct bioprospecting to identify novel enzymes with industrial potential, including halogenases for pharmaceutical synthesis and chitinases. We employ state-of-the-art annotation pipelines, combining homology-based searches with structural predictions via AlphaFold, to prioritize candidates for functional validation.
Working with colleagues in our NEXTMARINE project, we will express these enzymes in heterologous hosts like E. coli or yeast, optimizing conditions for high yield and stability, while assessing their activity through biochemical assays.
Image at right is a viral rhodopsin protein from Needham et al., PNAS 2019. Protein structure credit to Susumu Yoshizawa, Mikako Shirouzu, Wataru Iwasaki.

Use this space to share a testimonial quote about the business, its products or its services. Insert a quote from a real customer or client here to build trust and win over site visitors.
David M. Needham, Prof. Dr.
Principle Investigator


Teaching

Introduction to Biological Oceanography
MNF-bioc-101
This course surveys marine ecosystems from ocean physics and microbial processes to biogeochemical cycles and higher trophic levels across diverse habitats. Dr. Needham’s contributions are lectures on the Tree of Life and Viruses.
Kiel, Germany

Winter

Marine Microbiology
WASCAL-Microbio
This course, taught in Cabo Verde as part of the WASCAL Masters Program in 2022 and 2024, introduces the history, diversity, and ecological significance of marine microbes, emphasizing their roles in productivity, nutrient cycling, and biogeochemical processes. Students gain practical experience in microbial cultivation, microscopy, bioinformatics, and field sampling while applying ecological and molecular tools to understand microbial community structure and function.
Mindelo, Cabo Verde

Every two years

Microbial Ecology and Genomics
MNF-bioc-378
This course focuses on microbial ecology and genomics, emphasizing both conceptual understanding and hands-on experience with cutting-edge molecular tools and genomic data from the ocean. Through lectures, paper discussions, and practical exercises, students will explore microbial diversity, ecology, and dynamics while applying state-of-the-art techniques to unravel microbial roles in biogeochemical cycles.
Kiel, Germany

Winter

Scientific Computing and Bioinformatics
MNF-bioc-232
The course progresses from introductory skills in Bash, servers, and sequence quality analysis to advanced applications in rRNA gene analysis, metagenomics, and RNA-seq, combining statistical methods, visualization, and software installation along the way. Students gain experience across multiple molecular approaches, culminating in applications such as protein structure with GPUs.

Summer
River Watts
Arctic Ocean virus communities and their seasonality, bipolarity, and prokaryotic associations
River Watts
More realistic plankton simulation models will improve projections of ocean ecosystem responses to global change
River Watts
High-throughput, single-microbe genomics with strain resolution, applied to a human gut microbiome
River Watts
A distinct lineage of giant viruses brings a rhodopsin photosystem to unicellular marine predators
River Watts
Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples
River Watts
Metabolic and biogeochemical consequences of viral infection in aquatic ecosystems
River Watts
The microbiome of a bacterivorous marine choanoflagellate contains a resource-demanding obligate bacterial associate
River Watts
Basin-scale dynamics and enrichment-enabled genomics of marine nitrifiers: seasonality, niches, interac8ons, and genomic uniqueness
River Watts
From Microscale to Microbial Insights: Validating High-Throughput Microvolume Extraction Methods (HiMEx) for Microbial Ecology
Collaborations
Our international connections span multiple continents.
Some current and recent collaborators highlighted below.

Expedition

Global







